Working with output#
Utilities#
Several simply utilities exist to operate on output files
compare.py:this script takes two plot files and compares them zone-by-zone and reports the differences. This is useful for testing, to see if code changes affect the solution. Many problems have stored benchmarks in their solver’s tests directory. For example, to compare the current results for the incompressible shear problem to the stored benchmark, we would do:./compare.py shear_128_0216.pyro incompressible/tests/shear_128_0216.pyro
Differences on the order of machine precision may arise because of optimizations and compiler differences across platforms. Students should familiarize themselves with the details of how computers store numbers (floating point). An excellent read is What every computer scientist should know about floating-point arithmetic by D. Goldberg.
plot.py: this script uses the solver’sdovis()routine to plot an output file. For example, to plot the data in the fileshear_128_0216.pyrofrom the incompressible shear problem, you would do:./plot.py -o image.png shear_128_0216.pyro
where the
-ooption allows you to specify the output file name.
Reading and plotting manually#
pyro output data can be read using the util.io_pyro.read method. The following
sequence (done in a python session) reads in stored data (from the
compressible Sedov problem) and plots data falling on a line in the x
direction through the y-center of the domain. The return value of
read is a Simulation object.
import matplotlib.pyplot as plt
import pyro.util.io_pyro as io
sim = io.read("sedov_unsplit_0290.h5")
dens = sim.cc_data.get_var("density")
fig, ax = plt.subplots()
ax.plot(dens.g.x, dens[:,dens.g.qy//2])
ax.grid()
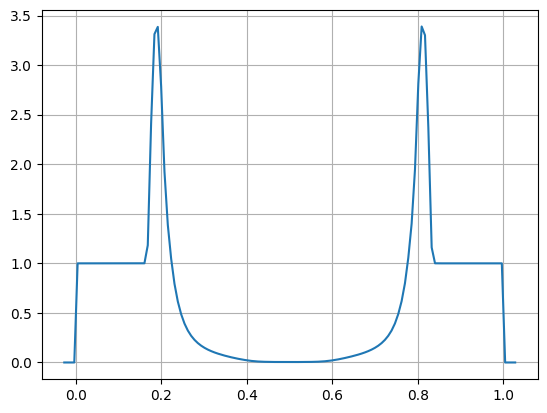
Note
This includes the ghost cells, by default, seen as the small
regions of zeros on the left and right. The total number of cells,
including ghost cells in the y-direction is qy, which is why
we use that in our slice.
If we wanted to exclude the ghost cells, then we could use the .v() method
on the density array to exclude the ghost cells, and then manually index g.x
to just include the valid part of the domain:
ax.plot(dens.g.x[g.ilo:g.ihi+1], dens.v()[:, dens.g.ny//2])
Note
In this case, we are using ny since that is the width of the domain
excluding ghost cells.
